The Impact of Common Gene Variants on EPA and DHA Status and Responsiveness to Increased Intakes
Professor Anne Marie Minihane
Department of Nutrition and Preventive Medicine, Norwich Medical School, University of East Anglia (UEA), Norwich, UK Website: https://www.uea.ac.uk/medicine/people/profile/a-minihane Setting the Scene A large body of human prospective epidemiology, as well as rodent feeding studies, have demonstrated the beneficial impact of the "marine" long chain n-3 PUFA (LC n-3 PUFA), EPA (20:5n-3) and DHA (22:6n-3), on fetal development, and cardiovascular and cognitive health. Randomised controlled trials (RCTs) have been less congruent, with this apparent lack of consistency likely attributable to a whole host of determinants of response, including habitual diet (including EPA and DHA intake and associated tissue status), medication use, the length of intervention, the total EPA+DHA dose and the EPA:DHA ratio, and the age, sex, BMI and health/disease status of the individual. In addition, it is likely that a large proportion of the inter-individual variability in response both within and between populations is attributable to common variants in genes involved in (i) modulating EPA and DHA status, including those central to EPA and DHA synthesis, transport, cell uptake and partitioning, and (ii) modulating their cellular and physiological targets, including transcription factor status (1). Although there is wide recognition that response to increased EPA+DHA intake is highly variable, currently generic dietary EPA and DHA recommendations are provided by international and national learned organisations such as the UK Scientific Advisory Committee in Nutrition (SACN), the International Society for the Study of Fatty Acids and Lipids (ISSFAL) and the American Heart Association (AHA). Typically a minimum dietary intake of 500 mg per day of EPA+DHA (achieved through the consumption of 1-2 portions of oily fish per week) is recommended for the general population, increasing to 1 g or 2-4 g per day for the secondary prevention of cardiovascular diseases (CVD) or as a TG lowering therapy, respectively. In the future, with the emergence of a fuller understanding of the aetiology of response, more stratified dietary guidelines may emerge, with recommended total EPA+DHA dose and EPA/DHA composition potentially based on a number of phenotypic and genotypic variables. Such a more targeted approach towards increased intake in responsive groups would also address the issue of marine n-3 PUFA sustainability. Current marine stocks provide only 40% of what is needed for all individuals globally to meet the conservative recommendation of > 500mg per day (2). Research providing an understanding of the genetic basis of the response to EPA and DHA intake and status is very much “work in progress.” Here, rather than be exhaustive, this invited opinion will focus on fatty acid desaturases (FADS), APOE, and PPAR genotypes, along with blood pressure and vascular function, as exemplars of variants in genes involved in EPA and DHA, synthesis, transport and cell uptake, transcription factor activation and a physiological target respectively, which may modulate EPA/DHA status and response to increased intakes. The Concise Temporal Guide to Genetics and Genotype and Its Assessment The origin of the science of genetics dates back to the 1850s-1860s, with Darwin and Wallace’s concept of Natural Selection and Gregor Mendel’s peas and the Laws of Inheritance. In 1953 Watson, Crick and Franklin described DNA as the molecule that carries genetic information and the unit of inheritance. But it was only in 2004 that the full sequence of the human genome (DNA) was described (3) and came the recognition that it contains 3 billion nucleotides (the structural unit consisting of a base, sugar and phosphate group) and far fewer genes (~22,000) than was originally thought. The genome sequence was considered by many to be the “panacea” in clinical medicine and public health, which would within a few years lead to considerable refinement in the prediction of risk of disease and the effective and wide-scale personalisation of preventive and therapeutic strategies. However, this was certainly an overly-optimistic speculation. The latest output from the 1000 Genome Consortium published in October 2015 (4) indicates that there are typically 88 million variants in a human genome. The identification of which variant predicts the progression from health to disease and response to environmental exposure, including altered food intake, will take a while to fully establish. Genetic variation comes in many “guises,” ranging from gross structural alterations (affecting more than 1000 bases/nucleotides in the DNA sequence) such as copy number variation, deletions, insertions and inversions, to single nucleotide polymorphisms (SNPs), where as the name suggests, a single base in a nucleotide is changed. Structural variants are relatively rare and often of high and largely un-modifiable penetrance (impact on health end-point). In contrast SNPs, which constitute > 90% of all genetic variability, are common and therefore most relevant for public health and will be the form of genetic variation considered here. Depending on the location of a SNP in the genome, it may have no functional impact or may affect gene expression (and therefore levels of proteins produced), or protein amino acid content, structure, stability and function. Approaches used to examine genotype*diet*phenotype associations can be targeted and hypothesis driven, with a focus on candidate genes with a known role in the metabolic process of interest. Publically available SNP databases such as the site curated by the National Centre for Biotechnology Information, US (www.ncbi.nlm.nih.gov/snp) are used to select SNPs of interest. In contrast non-targeted approaches, such as genome wide association studies (GWAS), which first appeared in 2005, may be used to identify novel loci associated with a phenotype of interest. In GWAS, the majority of the genome is assessed and genetic variation is compared between different sub-groups (such as those without or without disease, or responders and non-responders to intervention) to identify genotypes associated with a trait. How Variable is the Response to EPA and DHA Supplementation? In scientific publications data is almost exclusively presented as group means with some measure of variability in the form of e.g., SD or SEM, which disguises the large inter-individual variability in response. In the FINGEN study a total of n=312 healthy UK adults underwent a dose-response cross-over study consuming a control oil of fish oil providing 0.7 and 1.8g EPA+DHA per day in random order (5). A number of cardiovascular disease biomarkers were assessed including the plasma DHA and TG response. According to the records of returned capsules, compliance to treatment was 95% overall. In the group as a whole, a mean (SD) 11 (45) % and 67 (51) % decrease and increase in TG and DHA were evident following the high dose intervention for eight weeks, which represented ranges of -237% to 150% for TG and -137% to 253% for DHA (Figure 1) (5). Overall 118/312 participants had an increase in TG following intervention. 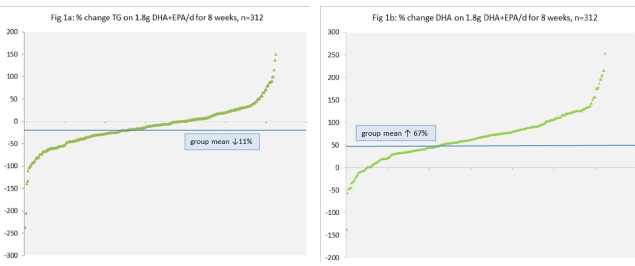 Fatty Acid Desaturase (FADS) Genotype, EPA/DHA Status and Health Endpoints Endogenous production of LC PUFAs from shorter-chain precursors (AA from LA, and EPA and DHA from αLNA) is mediated by the delta 5 (D5DS) and delta 6 desaturase (D6DS) enzymes, which are encoded by the FADS1 and FADS2 genes. In general, the efficiency rate for n-3 PUFAs is relatively low in humans, with an estimated conversion of αLNA to EPA of 0.2-6 % and <0.1% for DHA (6). It may be predicted that gene variants associated with greater D6DS and D5DS and activities would be associated with a higher tissue EPA and DHA status. This may be of particular relevance to individuals with a low consumption of fish who rely on plant derived αLNA to improve tissue EPA and DHA status. In a 2006 publication, which included an analysis of 18 SNPs of FADS1 and FADS2, the minor (less frequent) alleles were associated with higher levels of αLNA and LA and lower levels of DGLA, AA, EPA, and n-3 DPA, with no significant impact evident on DHA or n-6 DPA (7). These initial findings were broadly consistent with the output from the CHARGE consortium, which performed GWAS in five population-based cohorts comprising almost 9000 individuals of European ancestry (8). Associations between FADS1-FADS2 SNPs and haplotypes (grouping of SNPs) and blood lipids have also been observed using GWAS approaches (9). Furthermore, associations between variation in these gene loci and the incidence of diseases with a chronic inflammatory component have been reported (10), although Baylin et al. (11) failed to detect FADS SNP associations with incidence of myocardial infarction, despite genotype-dependent variation in tissue PUFA levels. In a fascinating recent GWAS analysis to investigate genetic signatures of diet and climate adaptation in Greenland Inuits (who have a high LC n-3 PUFA intake), FADS was the strongest loci associated with height, weight, growth hormone regulation and membrane fatty acid composition. In summary, individuals with a FADS genetic profile associated with low EPA and DHA synthesis may be particularly at risk from a lower tissue status and benefit from higher dietary intakes (12). APOE Genotype, EPA/DHA Status Lipids and Cognition Apolipoprotein E is an important lipid transporter in the circulation and in particular in the brain and acts as a high affinity ligand for cell uptake (13). Two common SNPs lead to three apoE isoforms, namely E2, E3 and E4. Individuals who carry an APOE4 allele, which is about 25% of Caucasians, are at significantly higher risk of dementia. Brain tissue is particularly enriched in DHA with concentrations 2-10 fold higher than in systemic tissues (14). The higher rate of cognitive decline in APOE4 carriers is likely to be in part due to a lower brain DHA status, defective uptake of DHA across the blood brain barrier (BBB) and increased DHA oxidation (15, 16). Lower enrichment in plasma DHA has been reported in APOE3/E4 individuals relative to the wild-type APOE3/E3 genotype (17) and the cognitive benefits of LC n-3 PUFA have been shown to be significantly less in APOE4 carriers relative to non-carriers (18).
Fatty Acid Desaturase (FADS) Genotype, EPA/DHA Status and Health Endpoints Endogenous production of LC PUFAs from shorter-chain precursors (AA from LA, and EPA and DHA from αLNA) is mediated by the delta 5 (D5DS) and delta 6 desaturase (D6DS) enzymes, which are encoded by the FADS1 and FADS2 genes. In general, the efficiency rate for n-3 PUFAs is relatively low in humans, with an estimated conversion of αLNA to EPA of 0.2-6 % and <0.1% for DHA (6). It may be predicted that gene variants associated with greater D6DS and D5DS and activities would be associated with a higher tissue EPA and DHA status. This may be of particular relevance to individuals with a low consumption of fish who rely on plant derived αLNA to improve tissue EPA and DHA status. In a 2006 publication, which included an analysis of 18 SNPs of FADS1 and FADS2, the minor (less frequent) alleles were associated with higher levels of αLNA and LA and lower levels of DGLA, AA, EPA, and n-3 DPA, with no significant impact evident on DHA or n-6 DPA (7). These initial findings were broadly consistent with the output from the CHARGE consortium, which performed GWAS in five population-based cohorts comprising almost 9000 individuals of European ancestry (8). Associations between FADS1-FADS2 SNPs and haplotypes (grouping of SNPs) and blood lipids have also been observed using GWAS approaches (9). Furthermore, associations between variation in these gene loci and the incidence of diseases with a chronic inflammatory component have been reported (10), although Baylin et al. (11) failed to detect FADS SNP associations with incidence of myocardial infarction, despite genotype-dependent variation in tissue PUFA levels. In a fascinating recent GWAS analysis to investigate genetic signatures of diet and climate adaptation in Greenland Inuits (who have a high LC n-3 PUFA intake), FADS was the strongest loci associated with height, weight, growth hormone regulation and membrane fatty acid composition. In summary, individuals with a FADS genetic profile associated with low EPA and DHA synthesis may be particularly at risk from a lower tissue status and benefit from higher dietary intakes (12). APOE Genotype, EPA/DHA Status Lipids and Cognition Apolipoprotein E is an important lipid transporter in the circulation and in particular in the brain and acts as a high affinity ligand for cell uptake (13). Two common SNPs lead to three apoE isoforms, namely E2, E3 and E4. Individuals who carry an APOE4 allele, which is about 25% of Caucasians, are at significantly higher risk of dementia. Brain tissue is particularly enriched in DHA with concentrations 2-10 fold higher than in systemic tissues (14). The higher rate of cognitive decline in APOE4 carriers is likely to be in part due to a lower brain DHA status, defective uptake of DHA across the blood brain barrier (BBB) and increased DHA oxidation (15, 16). Lower enrichment in plasma DHA has been reported in APOE3/E4 individuals relative to the wild-type APOE3/E3 genotype (17) and the cognitive benefits of LC n-3 PUFA have been shown to be significantly less in APOE4 carriers relative to non-carriers (18).
 In addition to influencing DHA metabolism and cognition, APOE genotype is known to influence the plasma lipid response to EPA+ DHA supplementation. A greater TG lowering effect has been observed in APOE4 carriers and in particular in males (5, 19), which may be due to a greater upregulation of lipoprotein lipase (LPL) (20), a key enzyme hydrolysing TG-rich lipoproteins (TRL) in the circulation. Therefore, APOE4 carriers represent a large ‘at-risk’ population subgroup who may particularly benefit from higher DHA intakes, although supra-physiological doses should be avoided in hyperlipidaemic individuals given that modest LDL-cholesterol raising effects have been observed (21). PPAR Genotype and the Plasma Lipid Response to EPA and DHA PPAR-α regulates multiple genes involved in TRL and HDL homeostasis such as LPL, apoA5, and apoA1 and in lipogenesis and fatty acid oxidation. It is the major transcription factor modulating the impact of EPA and DHA on plasma lipids. A number of publications have examined the impact of the PPAR-α Leu162Val variant, although findings have not been fully consistent (1). In the first of these analyses, based on the Framingham cohort, Val allele carriers had lower plasma TG and apoC3 concentrations relative to non-carriers, when consuming a high-PUFA diet, with n-6 PUFAs and n-3 PUFAs having comparable effects (22). No specific analysis of LC n-3 PUFAs was conducted. Rudkowska et al. indicated that the effect of PPAR-α Leu162Val genotype on the TG response to EPA+DHA supplementation in humans may be due to a differential impact on LPL activity (23). The authors went on to show in transfection studies that Leu162Leu homozygotes were more responsive to LC n-3 PUFAs with regard to LPL transcription (23). PPAR-γ regulates adipogenesis and lipid (TG) uptake and storage in adipose tissue. In vitro studies have shown that the Ala12 isoform of PPAR-γ2 is associated with a reduced ability to induce transcription and adipogenesis (24). In the KANWU Study carriers of the Ala12 allele had significantly greater reductions in serum TG levels in response to LC n-3 PUFA supplementation relative to the PPAR-γ2 Pro12Pro group (25). In a more recent publication, Jensen and co-workers reported that in addition to sex, this variant modulated the impact of FADS genotype on behaviour in children (26), highlighting the complexity of penetrance and interpretation of individual genotypes in isolation. Although not fully elucidated, given the central role PPARs play in modulating the effects of fatty acids on lipid metabolism, insulin sensitivity, vascular function, and inflammation, a research focus on this loci in the identification of the aetiology of the responsiveness of these physiological targets to EPA/DHA supplementation is merited. Genotype and the Variable Blood Pressure and Vascular Response to EPA and DHA Supplementation At a population level, it is thought that the hypotensive actions of EPA and DHA and their effect on vascular function and stiffness is only evident at high intakes, which could only be realistically achieved through high dose supplementation. However, it is likely that lower doses are effective in responsive individuals. In endothelial cells, endothelial nitric oxide synthase (eNOS) produces NO, a potent vasodilator. The eNOS Asp298Asp genotype has been associated with vascular function and cardiovascular risk (27) along with the vasodilatory response to EPA+DHA intake (28). Recent GWAS have identified a SNP in the c-Src tyrosine kinase (CSK) gene to be associated with blood pressure with an increase of systolic blood pressure of 0.6mmHg per allele (29). In the MARINA study CSK genotype emerged as a significant modulator of the systolic, diastolic and mean arterial blood pressure responses to EPA+DHA supplementation (30). Closing Remarks Although at a population level, dose-response relationships between EPA and DHA intakes and CVD clinical endpoints and biomarkers of CVD risk have been relatively quantitatively described, there is still a distinct lack of information on factors that determine individual responses. While progress is slower than originally expected, the published literature is providing insight into what factors may be important, with FADS and APOE genotypes emerging as important genetic modulators. Further identification of genetic determinants requires investigation in cohort studies where EPA and DHA intakes and status are accurately characterised, or in human RCTs that are sufficiently powered to study genotype*diet*phenotype associations. In the past small sample sizes are likely to be a major source of apparent lack of consistency between studies, and erroneous findings from such studies stunt progress in the field. Given the recognised biopotency of LC n-3 PUFAs along with evidence of considerable heterogeneity in response, current moves toward a more stratified approach to preventive and therapeutic medicine, and diminishing worldwide marine stocks, investigations in this area are of wide public health relevance. However, we must not get too “gene-centric” in our thinking and research focus, and recognise that effective future personalisation must consider other major factors that are likely to influence response to EPA and DHA such as sex, habitual intake and disease status. References 1. Madden J, Williams CM, Calder PC, Lietz G, Miles EA, Cordell H, Mathers JC, Minihane AM. The impact of common gene variants on the response of biomarkers of cardiovascular disease (CVD) risk to increased fish oil fatty acids intakes. Annual Review of Nutrition 2011;31:203-34. [link] 2. FAO. The State of World Fisheries and Aquaculture 2012. 2012. [link] 3. Lander ES. Initial impact of the sequencing of the human genome. Nature 2011;470(7333):187-97. [link] 4. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015;526(7571):68-74. [link] 5. Caslake MJ, Miles EA, Kofler BM, Lietz G, Curtis P, Armah CK, Kimber AC, Grew JP, Farrell L, Stannard J, et al. Effect of sex and genotype on cardiovascular biomarker response to fish oils: the FINGEN Study. The American Journal of Clinical Nutrition 2008;88(3):618-29. [link] 6. Burdge GC. Metabolism of alpha-linolenic acid in humans. Prostaglandins, Leukotrienes, and Essential Fatty Acids 2006;75(3):161-8. [link] 7. Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, Illig T, Koletzko B, Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Human Molecular Genetics 2006;15(11):1745-56. [link] 8. Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng LC, Bhattacharya S, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genetics 2011;7(7):e1002193. [link] 9. Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nature Genetics 2009;41(1):47-55. [link] 10. Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, Sandri M, Friso S, Pizzolo F, Schaeffer L, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. The American Journal of Clinical Nutrition 2008;88(4):941-9. [link] 11. Baylin A, Ruiz-Narvaez E, Kraft P, Campos H. alpha-Linolenic acid, Delta6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. The American Journal of Clinical Nutrition 2007;85(2):554-60. [link] 12. Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science (New York, NY) 2015;349(6254):1343-7. [link] 13. Rimbach G, Minihane AM. Nutrigenetics and personalised nutrition: how far have we progressed and are we likely to get there? The Proceedings of the Nutrition Society 2009;68(2):162-72. [link] 14. Crawford MA, Bloom M, Broadhurst CL, Schmidt WF, Cunnane SC, Galli C, Gehbremeskel K, Linseisen F, Lloyd-Smith J, Parkington J. Evidence for the unique function of docosahexaenoic acid during the evolution of the modern hominid brain. Lipids 1999;34 Suppl:S39-47. [link] 15. Chouinard-Watkins R, Rioux-Perreault C, Fortier M, Tremblay-Mercier J, Zhang Y, Lawrence P, Vohl MC, Perron P, Lorrain D, Brenna JT, et al. Disturbance in uniformly 13C-labelled DHA metabolism in elderly human subjects carrying the apoE epsilon4 allele. The British Journal of Nutrition 2013;110(10):1751-9. [link] 16. Vandal M, Alata W, Tremblay C, Rioux-Perreault C, Salem N, Jr., Calon F, Plourde M. Reduction in DHA transport to the brain of mice expressing human APOE4 compared to APOE2. Journal of Neurochemistry 2014;129(3):516-26. [link] 17. Chouinard-Watkins R, Conway V, Minihane AM, Jackson KG, Lovegrove JA, Plourde M. Interaction between BMI and APOE genotype is associated with changes in the plasma long-chain-PUFA response to a fish-oil supplement in healthy participants. The American Journal of Clinical Nutrition 2015;102(2):505-13. [link] 18. Huang TL, Zandi PP, Tucker KL, Fitzpatrick AL, Kuller LH, Fried LP, Burke GL, Carlson MC. Benefits of fatty fish on dementia risk are stronger for those without APOE epsilon4. Neurology 2005;65(9):1409-14. [link] 19. Carvalho-Wells AL, Jackson KG, Lockyer S, Lovegrove JA, Minihane AM. APOE genotype influences triglyceride and C-reactive protein responses to altered dietary fat intake in UK adults. The American Journal of Clinical Nutrition 2012;96(6):1447-53. [link] 20. Khan S, Minihane AM, Talmud PJ, Wright JW, Murphy MC, Williams CM, Griffin BA. Dietary long-chain n-3 PUFAs increase LPL gene expression in adipose tissue of subjects with an atherogenic lipoprotein phenotype. Journal of Lipid Research 2002;43(6):979-85. [link] 21. Olano-Martin E, Anil E, Caslake MJ, Packard CJ, Bedford D, Stewart G, Peiris D, Williams CM, Minihane AM. Contribution of apolipoprotein E genotype and docosahexaenoic acid to the LDL-cholesterol response to fish oil. Atherosclerosis 2010;209(1):104-10. [link] 22. Tai ES, Corella D, Demissie S, Cupples LA, Coltell O, Schaefer EJ, Tucker KL, Ordovas JM. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. The Journal of Nutrition 2005;135(3):397-403. [link] 23. Rudkowska I, Caron-Dorval D, Verreault M, Couture P, Deshaies Y, Barbier O, Vohl MC. PPARalpha L162V polymorphism alters the potential of n-3 fatty acids to increase lipoprotein lipase activity. Molecular Nutrition & Food Research 2010;54(4):543-50. [link] 24. Deeb SS, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics 1998;20(3):284-7. [link] 25. Lindi V, Schwab U, Louheranta A, Laakso M, Vessby B, Hermansen K, Storlien L, Riccardi G, A AR. Impact of the Pro12Ala polymorphism of the PPAR-gamma2 gene on serum triacylglycerol response to n-3 fatty acid supplementation. Molecular Genetics and Metabolism 2003;79(1):52-60. [link] 26. Jensen HA, Harslof LB, Nielsen MS, Christensen LB, Ritz C, Michaelsen KF, Vogel U, Lauritzen L. FADS single-nucleotide polymorphisms are associated with behavioral outcomes in children, and the effect varies between sexes and is dependent on PPAR genotype. The American Journal of Clinical Nutrition 2014;100(3):826-32. [link] 27. Li J, Wu X, Li X, Feng G, He L, Shi Y. The endothelial nitric oxide synthase gene is associated with coronary artery disease: a meta-analysis. Cardiology 2010;116(4):271-8. [link] 28. Thompson AK, Newens KJ, Jackson KG, Wright J, Williams CM. Glu298Asp polymorphism influences the beneficial effects of fish oil fatty acids on postprandial vascular function. Journal of Lipid Research 2012;53(10):2205-13. [link] 29. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478(7367):103-9. [link] 30. AlSaleh A, Maniou Z, Lewis FJ, Hall WL, Sanders TA, O'Dell SD. Interaction between a CSK gene variant and fish oil intake influences blood pressure in healthy adults. The Journal of Nutrition 2014;144(3):267-72. [link]
In addition to influencing DHA metabolism and cognition, APOE genotype is known to influence the plasma lipid response to EPA+ DHA supplementation. A greater TG lowering effect has been observed in APOE4 carriers and in particular in males (5, 19), which may be due to a greater upregulation of lipoprotein lipase (LPL) (20), a key enzyme hydrolysing TG-rich lipoproteins (TRL) in the circulation. Therefore, APOE4 carriers represent a large ‘at-risk’ population subgroup who may particularly benefit from higher DHA intakes, although supra-physiological doses should be avoided in hyperlipidaemic individuals given that modest LDL-cholesterol raising effects have been observed (21). PPAR Genotype and the Plasma Lipid Response to EPA and DHA PPAR-α regulates multiple genes involved in TRL and HDL homeostasis such as LPL, apoA5, and apoA1 and in lipogenesis and fatty acid oxidation. It is the major transcription factor modulating the impact of EPA and DHA on plasma lipids. A number of publications have examined the impact of the PPAR-α Leu162Val variant, although findings have not been fully consistent (1). In the first of these analyses, based on the Framingham cohort, Val allele carriers had lower plasma TG and apoC3 concentrations relative to non-carriers, when consuming a high-PUFA diet, with n-6 PUFAs and n-3 PUFAs having comparable effects (22). No specific analysis of LC n-3 PUFAs was conducted. Rudkowska et al. indicated that the effect of PPAR-α Leu162Val genotype on the TG response to EPA+DHA supplementation in humans may be due to a differential impact on LPL activity (23). The authors went on to show in transfection studies that Leu162Leu homozygotes were more responsive to LC n-3 PUFAs with regard to LPL transcription (23). PPAR-γ regulates adipogenesis and lipid (TG) uptake and storage in adipose tissue. In vitro studies have shown that the Ala12 isoform of PPAR-γ2 is associated with a reduced ability to induce transcription and adipogenesis (24). In the KANWU Study carriers of the Ala12 allele had significantly greater reductions in serum TG levels in response to LC n-3 PUFA supplementation relative to the PPAR-γ2 Pro12Pro group (25). In a more recent publication, Jensen and co-workers reported that in addition to sex, this variant modulated the impact of FADS genotype on behaviour in children (26), highlighting the complexity of penetrance and interpretation of individual genotypes in isolation. Although not fully elucidated, given the central role PPARs play in modulating the effects of fatty acids on lipid metabolism, insulin sensitivity, vascular function, and inflammation, a research focus on this loci in the identification of the aetiology of the responsiveness of these physiological targets to EPA/DHA supplementation is merited. Genotype and the Variable Blood Pressure and Vascular Response to EPA and DHA Supplementation At a population level, it is thought that the hypotensive actions of EPA and DHA and their effect on vascular function and stiffness is only evident at high intakes, which could only be realistically achieved through high dose supplementation. However, it is likely that lower doses are effective in responsive individuals. In endothelial cells, endothelial nitric oxide synthase (eNOS) produces NO, a potent vasodilator. The eNOS Asp298Asp genotype has been associated with vascular function and cardiovascular risk (27) along with the vasodilatory response to EPA+DHA intake (28). Recent GWAS have identified a SNP in the c-Src tyrosine kinase (CSK) gene to be associated with blood pressure with an increase of systolic blood pressure of 0.6mmHg per allele (29). In the MARINA study CSK genotype emerged as a significant modulator of the systolic, diastolic and mean arterial blood pressure responses to EPA+DHA supplementation (30). Closing Remarks Although at a population level, dose-response relationships between EPA and DHA intakes and CVD clinical endpoints and biomarkers of CVD risk have been relatively quantitatively described, there is still a distinct lack of information on factors that determine individual responses. While progress is slower than originally expected, the published literature is providing insight into what factors may be important, with FADS and APOE genotypes emerging as important genetic modulators. Further identification of genetic determinants requires investigation in cohort studies where EPA and DHA intakes and status are accurately characterised, or in human RCTs that are sufficiently powered to study genotype*diet*phenotype associations. In the past small sample sizes are likely to be a major source of apparent lack of consistency between studies, and erroneous findings from such studies stunt progress in the field. Given the recognised biopotency of LC n-3 PUFAs along with evidence of considerable heterogeneity in response, current moves toward a more stratified approach to preventive and therapeutic medicine, and diminishing worldwide marine stocks, investigations in this area are of wide public health relevance. However, we must not get too “gene-centric” in our thinking and research focus, and recognise that effective future personalisation must consider other major factors that are likely to influence response to EPA and DHA such as sex, habitual intake and disease status. References 1. Madden J, Williams CM, Calder PC, Lietz G, Miles EA, Cordell H, Mathers JC, Minihane AM. The impact of common gene variants on the response of biomarkers of cardiovascular disease (CVD) risk to increased fish oil fatty acids intakes. Annual Review of Nutrition 2011;31:203-34. [link] 2. FAO. The State of World Fisheries and Aquaculture 2012. 2012. [link] 3. Lander ES. Initial impact of the sequencing of the human genome. Nature 2011;470(7333):187-97. [link] 4. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015;526(7571):68-74. [link] 5. Caslake MJ, Miles EA, Kofler BM, Lietz G, Curtis P, Armah CK, Kimber AC, Grew JP, Farrell L, Stannard J, et al. Effect of sex and genotype on cardiovascular biomarker response to fish oils: the FINGEN Study. The American Journal of Clinical Nutrition 2008;88(3):618-29. [link] 6. Burdge GC. Metabolism of alpha-linolenic acid in humans. Prostaglandins, Leukotrienes, and Essential Fatty Acids 2006;75(3):161-8. [link] 7. Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, Illig T, Koletzko B, Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Human Molecular Genetics 2006;15(11):1745-56. [link] 8. Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng LC, Bhattacharya S, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genetics 2011;7(7):e1002193. [link] 9. Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nature Genetics 2009;41(1):47-55. [link] 10. Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, Sandri M, Friso S, Pizzolo F, Schaeffer L, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. The American Journal of Clinical Nutrition 2008;88(4):941-9. [link] 11. Baylin A, Ruiz-Narvaez E, Kraft P, Campos H. alpha-Linolenic acid, Delta6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. The American Journal of Clinical Nutrition 2007;85(2):554-60. [link] 12. Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science (New York, NY) 2015;349(6254):1343-7. [link] 13. Rimbach G, Minihane AM. Nutrigenetics and personalised nutrition: how far have we progressed and are we likely to get there? The Proceedings of the Nutrition Society 2009;68(2):162-72. [link] 14. Crawford MA, Bloom M, Broadhurst CL, Schmidt WF, Cunnane SC, Galli C, Gehbremeskel K, Linseisen F, Lloyd-Smith J, Parkington J. Evidence for the unique function of docosahexaenoic acid during the evolution of the modern hominid brain. Lipids 1999;34 Suppl:S39-47. [link] 15. Chouinard-Watkins R, Rioux-Perreault C, Fortier M, Tremblay-Mercier J, Zhang Y, Lawrence P, Vohl MC, Perron P, Lorrain D, Brenna JT, et al. Disturbance in uniformly 13C-labelled DHA metabolism in elderly human subjects carrying the apoE epsilon4 allele. The British Journal of Nutrition 2013;110(10):1751-9. [link] 16. Vandal M, Alata W, Tremblay C, Rioux-Perreault C, Salem N, Jr., Calon F, Plourde M. Reduction in DHA transport to the brain of mice expressing human APOE4 compared to APOE2. Journal of Neurochemistry 2014;129(3):516-26. [link] 17. Chouinard-Watkins R, Conway V, Minihane AM, Jackson KG, Lovegrove JA, Plourde M. Interaction between BMI and APOE genotype is associated with changes in the plasma long-chain-PUFA response to a fish-oil supplement in healthy participants. The American Journal of Clinical Nutrition 2015;102(2):505-13. [link] 18. Huang TL, Zandi PP, Tucker KL, Fitzpatrick AL, Kuller LH, Fried LP, Burke GL, Carlson MC. Benefits of fatty fish on dementia risk are stronger for those without APOE epsilon4. Neurology 2005;65(9):1409-14. [link] 19. Carvalho-Wells AL, Jackson KG, Lockyer S, Lovegrove JA, Minihane AM. APOE genotype influences triglyceride and C-reactive protein responses to altered dietary fat intake in UK adults. The American Journal of Clinical Nutrition 2012;96(6):1447-53. [link] 20. Khan S, Minihane AM, Talmud PJ, Wright JW, Murphy MC, Williams CM, Griffin BA. Dietary long-chain n-3 PUFAs increase LPL gene expression in adipose tissue of subjects with an atherogenic lipoprotein phenotype. Journal of Lipid Research 2002;43(6):979-85. [link] 21. Olano-Martin E, Anil E, Caslake MJ, Packard CJ, Bedford D, Stewart G, Peiris D, Williams CM, Minihane AM. Contribution of apolipoprotein E genotype and docosahexaenoic acid to the LDL-cholesterol response to fish oil. Atherosclerosis 2010;209(1):104-10. [link] 22. Tai ES, Corella D, Demissie S, Cupples LA, Coltell O, Schaefer EJ, Tucker KL, Ordovas JM. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. The Journal of Nutrition 2005;135(3):397-403. [link] 23. Rudkowska I, Caron-Dorval D, Verreault M, Couture P, Deshaies Y, Barbier O, Vohl MC. PPARalpha L162V polymorphism alters the potential of n-3 fatty acids to increase lipoprotein lipase activity. Molecular Nutrition & Food Research 2010;54(4):543-50. [link] 24. Deeb SS, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics 1998;20(3):284-7. [link] 25. Lindi V, Schwab U, Louheranta A, Laakso M, Vessby B, Hermansen K, Storlien L, Riccardi G, A AR. Impact of the Pro12Ala polymorphism of the PPAR-gamma2 gene on serum triacylglycerol response to n-3 fatty acid supplementation. Molecular Genetics and Metabolism 2003;79(1):52-60. [link] 26. Jensen HA, Harslof LB, Nielsen MS, Christensen LB, Ritz C, Michaelsen KF, Vogel U, Lauritzen L. FADS single-nucleotide polymorphisms are associated with behavioral outcomes in children, and the effect varies between sexes and is dependent on PPAR genotype. The American Journal of Clinical Nutrition 2014;100(3):826-32. [link] 27. Li J, Wu X, Li X, Feng G, He L, Shi Y. The endothelial nitric oxide synthase gene is associated with coronary artery disease: a meta-analysis. Cardiology 2010;116(4):271-8. [link] 28. Thompson AK, Newens KJ, Jackson KG, Wright J, Williams CM. Glu298Asp polymorphism influences the beneficial effects of fish oil fatty acids on postprandial vascular function. Journal of Lipid Research 2012;53(10):2205-13. [link] 29. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478(7367):103-9. [link] 30. AlSaleh A, Maniou Z, Lewis FJ, Hall WL, Sanders TA, O'Dell SD. Interaction between a CSK gene variant and fish oil intake influences blood pressure in healthy adults. The Journal of Nutrition 2014;144(3):267-72. [link]

